Graduate Student Openings, Fall 2024
There are multiple openings in the group for Fall 2024 in the different sub-areas within our group. Students could also choose the projects that fall at the interface between two research areas. Students will also have the freedom to shape their projects. Our current graduate students are from the departments of Chemical and Biomolecular Engineering, Chemistry, Bioengineering, Biophysics and Information Sciences. In particular, we are planning to recruit students for the following projects.
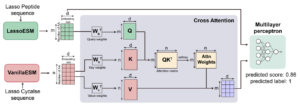
Project 1. AI based methods for design of Ribosomally synthesized and post-translationally modified peptides (1 Machine Learning + Molecular Dynamics Position). RiPPs are structurally diverse bioactive natural products. RiPPs biosynthetic enzymes can be extremely substrate tolerant, which makes it possible to engineer novel compounds for biomedical applications. However, predicting enzyme substrate tolerance and properties like bioactivity remains challenging due to limited experimental data and the intricate nature of the substrate fitness landscapes. In this project, we will leverage high-throughput experiments on biosynthetic enzymes involved in making RIPPs along with AI based methods to understand the design rules for the origin of substrate specificity. We have already developed several large language models for peptides, lasso peptides and protein-peptide interactions that would be deployed in this project as well. This project is supported by National Institutes of Health grant in collaboration with Prof. Wilfred Van der Donk (Chemistry), Doug Mitchell (Chemistry) and Huimin Zhao (ChBE).
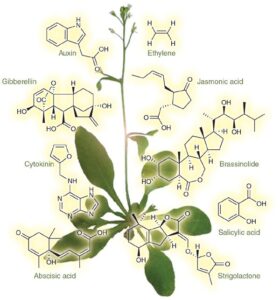 Project 2. Large Language Models for Design of Agrochemicals ( 1 Machine Learning + Experimental Position). Plant hormones are small molecules that regulate plant growth, development, and responses to biotic and abiotic stresses. They are specifically recognized by the binding site of their receptors. In this work, we employ experiments and advanced AI methods for design of molecules to mimic the eight classes of phytohormones (auxin, jasmonate, gibberellin, strigolactone, brassinosteroid, cytokinin, salicylic acid, and abscisic acid) to their canonical receptors. The student will not only develop large language models specific to agrochemicals but they will also have a chance to perfrom experiments to validate the molecules designed by generative AI. This project is supported by the Frasch Foundation.
Project 2. Large Language Models for Design of Agrochemicals ( 1 Machine Learning + Experimental Position). Plant hormones are small molecules that regulate plant growth, development, and responses to biotic and abiotic stresses. They are specifically recognized by the binding site of their receptors. In this work, we employ experiments and advanced AI methods for design of molecules to mimic the eight classes of phytohormones (auxin, jasmonate, gibberellin, strigolactone, brassinosteroid, cytokinin, salicylic acid, and abscisic acid) to their canonical receptors. The student will not only develop large language models specific to agrochemicals but they will also have a chance to perfrom experiments to validate the molecules designed by generative AI. This project is supported by the Frasch Foundation.
 Project 3. High-throughput characterization of membrane proteins using deep mutational scans (1 Experimental+Molecular Simulations Position). Deep mutagenesis, whereby tens of thousands of mutational effects are determined by combining in vitro selections of sequence variants with deep sequencing. The overall aim of the research is to use deep mutagenesis and molecular dynamics simulations synergistically to determine the mechanism of membrane proteins such as ion-coupled neurotransmitter transporters, Sugar transporters and G-protein coupled receptors (GPCRs). We plan to recruit two students working on experimental and computational aspects of this projects. This research project is supported by the National Institutes of Health.
Project 3. High-throughput characterization of membrane proteins using deep mutational scans (1 Experimental+Molecular Simulations Position). Deep mutagenesis, whereby tens of thousands of mutational effects are determined by combining in vitro selections of sequence variants with deep sequencing. The overall aim of the research is to use deep mutagenesis and molecular dynamics simulations synergistically to determine the mechanism of membrane proteins such as ion-coupled neurotransmitter transporters, Sugar transporters and G-protein coupled receptors (GPCRs). We plan to recruit two students working on experimental and computational aspects of this projects. This research project is supported by the National Institutes of Health.
Project 4. Deep learning models for plant enzyme engineering (2 Machine Learning + Experimental position). Engineering of plant enzyme variants requires strategies for sequence design based on the measured enzyme properties. Experimental enzyme properties such as Michalis constant (Km) and catalytic efficiency (kcat/Km) have been documented in the BRENDA and Sabio-RK databases. However, the data on plant enzymes in public datasets is limited. Experimental measurements of these properties are challenging and labor-intensive. Therefore, there is a need for an in silico predictor that could provide a rapid estimate of enzyme properties based on sequence. We aim to develop plant enzyme specific deep learning models for property prediction by augmenting the enzyme properties databases with both computational and experimental information about plant enzymes. This work is supported by National Science Foundation and is pursued in collaboration with Prof. Andrew Leakey (Plant Biology) and Prof. Matthew Hudson (Crop Sciences).
Postdoctoral Fellow (Experimental), Fall 2024
We are seeking a talented Postdoctoral Associate to join our experimental team. The ideal candidate will possess excellent academic credentials in experimental molecular biology, as well as expertise or knowledge in chemical biology.
In this role, the successful applicant will be a key member within the laboratory and make significant contributions to the advancement of group’s protein engineering research with emphasis on engineering proteins in plants. Primarily, the candidate will be responsible for the conducting high-throughput experiments for either generating protein sequence-function data or high-throughput screening of ligands to enhance the potency and selectivity of potential drug candidates. Furthermore, the candidate will enjoy ample opportunities to learn and actively participate in multidisciplinary projects in our group, making a meaningful impact and engaging in independent ideation to drive the group’s research. We are also looking for candidates who are interested in learning the basics of AI and ML and apply these ideas in their project. There will be many opportunities for collaboration with other experimental groups as well.
The position presents an exciting and fulfilling prospect for career growth, as it involves pioneering work in the field of protein engineering and also provides ample opportunities for learning new skills.

