Predicting the Activities of Drug Excipients on Biological Targets using One-Shot Learning
BibTeX
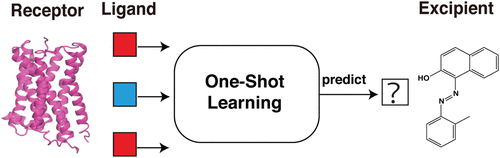
Excipients are a major component of drugs and are used to improve drugs attributes such as stability and appearance. Excipients approved by Food and Drug Administration (FDA) are regarded as safe for human in allowed concentration, but their potential interaction with drug targets have not been investigated systematically, which might influence drug’s efficacy. Deep learning models have been used for identification of ligands that could bind to the drug targets. However, due to the limited available data, it is challenging to reliably estimate the likelihood of a ligand-protein interaction. One-shot learning techniques provide a potential approach to address this low-data problem as these techniques require only one or a few examples to classify the new data. In this study, we apply one-shot learning models on datasets that include ligands binding to G-Protein Coupled Receptors (GPCRs) and Kinases. The predicted results suggest that one-shot learning models could be used for predicting ligand-protein interaction and the models attain better performance when protein targets contain conserved binding pockets. The trained models are also used to predict interactions between excipients and drug targets, which provides a potential efficient strategy to explore the activities of drug excipients. We find that a large number of drug excipients could interact with biological targets and influence their function. The results demonstrate how one-shot learning models can be used to make accurate prediction for excipient-protein interactions and these methods could be used for selecting excipients with limited drug-protein interactions.

